Populations » Quality Control in AFND
Introduction
AFND is a public resource that collects information on allele, genotype and haplotype frequencies from different polymorphic areas in the human genome such as human leukocyte antigens (HLA), killer-cell immunoglobulin-like receptors, etc. To produce this database we have compiled a large collection of datasets from different sources including: (i) peer-reviewed literature, (ii) datasets from international workshops in immunogenetics and histocompatiblity and (iii) data submitted directly to AFND by individual laboratories. As more than 75% of the submissions in AFND are derived from peer-review literature, we rely upon data verification by journal editors and reviewers when source studies are published. However, in an effort to (re)assess the data, curators of AFND apply several rules to check the accuracy of the data as shown in Figure 1.
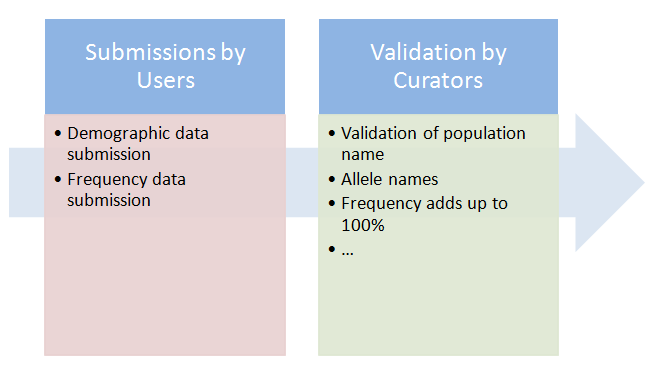 |
In the following sections, we describe several reports to explain how the data is submitted and validated in AFND. If you have any query please do not hesitate to contact us.
1 Validation of demographics data
1.1 Population names
ο How we name a population? | |
|
Each population in AFND is named according to the combination of the country name, geographical location
and ethnic group when available, to describe the population in as much detail as possible. E.g. South Africa Natal Zulu In order to identify the polymorphic region studied in a given population an additional word is incorporated at the end of the name, except for HLA populations which were the first populations entered in the database. E.g. South Africa Xhosa KIR, Italy Milan Cytokine, etc. If another set of individuals from a given population, which was geographically and ethnically similar to an existing population in the database, was submitted, a consecutive number was assigned to that population to differentiate the two populations. E.g. China Guangzhou Han, China Guangzhou Han pop 2 In some populations, individuals are living or born in a different country from their original ethnic background (i.e., immigrants from a different country). In these cases, the name of the original ethnic background was included in the name of the population, and the country was defined as the current location in which individuals were living/born. For these populations, users can search data using either country as filter. E.g. Germany DKMS Turkey minority (Turkish descendant people living in Germany), Singapore Thai (Thai descendant people living in Singapore) Finally, many countries may have populations typed from different sources. Thus, if users are interested in populations from one country, we recommend that the search is performed for populations from that country initially, then users can filter populations according to specific sources. E.g. Anthropology. | |
ο How are the geographical regions assigned to each population in AFND? | |
|
In AFND, we have organised the populations by geographical regions. In collaboration with the IDAWG and HLA-NET consortiums,
we have defined 12 geographical regions.
Tool available: Click here to consult the geographical regions. | |
2 Validation of frequency data for HLA
2.1 Allele names
ο Why an HLA allele has been changed in AFND compared to the original publication? | |
| Allele names may have been changed in the IMGT/HLA database (Official Nomenclature Database) after their original submission for different reasons. For example, the sequence of the allele A*01:34N was shown to be expressed at low levels and the allele was renamed to A*01:01:38L in March 2011. Thus, in AFND, we have inputted the allele frequency under the new allele code. | |
ο Is serology-based nomenclature data inputted in AFND? | |
| Some populations were typed only by serology. In these instances, we have converted data into the IMGT/HLA database nomenclature. | |
ο How AFND handles data from low and high resolution alleles? | |
| AFND collects data at different level of resolution. We automatically generate all possible low resolution allele names based on the IMGT/HLA catalogue. For example, the allele A*01:01:01:01 is automatically split to generate frequencies for A*01, A*01:01 and A*01:01:01 alleles. | |
ο How is AFND updated with the new nomenclature from IMGT/HLA? | |
| AFND receives quarterly reports from the IMGT/HLA, i.e. every new release. We update AFND according to the latest
release immediately after this notification.
Tool available: Click here to consult the latest IMGT/HLA release in AFND. | |
2.2 Frequencies
ο Why some HLA alleles have frequencies > 50%? | |
| Based on the high diversity of HLA alleles, one may expect that the frequencies of alleles should not exceed 50%.
However, we have detected some populations which allele frequencies are over 50%. This may be the case of certain loci that have a low number of alleles, such as
DPA1, DQB1, etc., or in some populations that have very few alleles at a given locus.
We have also examined those populations that have > 75% of the individuals with that allele.
Tool available: Click here to consult the list of populations which have alleles with frequencies over 50% or the percentage of individuals > 75%. | |
ο Why some populations have allele frequencies that do not add up to 100%? | |
| In some cases, authors have excluded in their publications those alleles reported at low frequencies.
To specify this, we have included a sentence in the demographics of the population.
Tool available: Click here to consult the list of populations which do not add up to 100% for allele frequencies. | |
ο Why some populations have haplotype frequencies that do not add up to 100%? | |
| For some populations all haplotypes are not necessarily listed. Sometimes this is
because not all haplotypes are listed in the publication because they are at a very low frequency. As far as possible, we have
listed all haplotypes greater than 1%. When the population is large, we have added haplotypes at lower percentages.
Tool available: Click here to consult the list of populations which do not add up to 100% for haplotype frequencies. | |
ο Why some populations have both low and high resolution data? | |
| We decided to capture both allele frequencies at high and low resolution, by summing high resolution data to produce low resolution frequency data.
For example, the Northern Ireland population in AFND will have frequencies for A*25:01 = 0.0200, A*25:02 = 0.0010 and the sum of these two A*25 = 0.0210.
Tool available: Click here to consult the list of populations that have both, low and high resolution frequencies. | |
ο Why some alleles have frequencies shown as 0.000 and some have frequencies with no data? | |
| If it is known that an allele/gene has been tested for and not found, we have entered 0.000. If the allele/gene has not been tested for, the frequency column is left in blank. | |
ο Why the allele frequencies differ from counts/sample size? | |
| In some cases, the number individuals sampled (typed) for each locus within the same population are different. Thus, users are advised to check demographic data of the pops they have searched for to ascertain possible deviations in data. In instances, where alleles could not be distinguished, we would put the frequency under the first allele but adding a note in demographic information. | |
3 Validation of frequency data for KIR
3.1 Allele names
ο How is AFND updated with the new nomenclature from IPD-KIR? | |
| AFND receives reports from the IPD-KIR every new release. We update AFND according to the latest
release immediately after this notification.
Tool available: Click here to consult the latest IPD-KIR release in AFND | |
3.2 Frequencies
ο Why some KIR populations have genotype frequencies that do not add up to 100%? | |
| In some cases, some authors have only reported the most common KIR genotypes found in a given population.
Thus, some populations may not add up to 100% if we add all genotype frequencies.
Tool available: Click here to consult those populations that do not add up to 100% | |